Embedding¶
Canonical Correlation Analysis (CCA)¶
- class mvlearn.embed.CCA(n_components=1, regs=None, signal_ranks=None, center=True, i_mcca_method='auto', multiview_output=True)[source]¶
Canonical Correlation Analysis (CCA)
CCA inherits from MultiCCA (MCCA) but is restricted to 2 views which allows for certain statistics to be computed about the results.
- Parameters
n_components (int (default 1)) -- Number of canonical components to compute and return.
regs (float | 'lw' | 'oas' | None, or list, optional (default None)) --
CCA regularization for each data view, which can be important for high dimensional data. A list will specify for each view separately. If float, must be between 0 and 1 (inclusive).
0 or None : corresponds to SUMCORR-AVGVAR MCCA.
1 : partial least squares SVD (generalizes to more than 2 views)
'lw' : Default
sklearn.covariance.ledoit_wolfregularization'oas' : Default
sklearn.covariance.oasregularization
signal_ranks (int, None or list, optional (default None)) -- The initial signal rank to compute. If None, will compute the full SVD. A list will specify for each view separately.
center (bool, or list (default True)) -- Whether or not to initially mean center the data. A list will specify for each view separately.
i_mcca_method ('auto' | 'svd' | 'gevp' (default 'auto')) -- Whether or not to use the SVD based method (only works with no regularization) or the gevp based method for informative MCCA.
multiview_output (bool, optional (default True)) -- If True, the
.transformmethod returns one dataset per view. Otherwise, it returns one dataset, of shape (n_samples, n_components)
- means_¶
The means of each view, each of shape (n_features,)
- Type
list of numpy.ndarray
- loadings_¶
The loadings for each view used to project new data, each of shape (n_features_b, n_components).
- Type
list of numpy.ndarray
- common_score_norms_¶
Column norms of the sum of the fitted view scores. Used for projecting new data
- Type
numpy.ndarray, shape (n_components,)
- evals_¶
The generalized eigenvalue problem eigenvalues.
- Type
numpy.ndarray, shape (n_components,)
References
- 1
Kettenring, J. R., "Canonical Analysis of Several Sets of Variables." Biometrika, 58:433-451, (1971)
- 2
Tenenhaus, A., et al. "Regularized generalized canonical correlation analysis." Psychometrika, 76:257–284, 2011
Examples
>>> from mvlearn.embed import CCA >>> X1 = [[0., 0., 1.], [1.,0.,0.], [2.,2.,2.], [3.,5.,4.]] >>> X2 = [[0.1, -0.2], [0.9, 1.1], [6.2, 5.9], [11.9, 12.3]] >>> cca = CCA() >>> cca.fit([X1, X2]) CCA() >>> Xs_scores = cca.transform([X1, X2])
Multiview Canonical Correlation Analysis (MCCA)¶
- class mvlearn.embed.MCCA(n_components=1, regs=None, signal_ranks=None, center=True, i_mcca_method='auto', multiview_output=True)[source]¶
Multiview canonical correlation analysis for any number of views. Includes options for regularized MCCA and informative MCCA (where a low rank PCA is first computed).
- Parameters
n_components (int | 'min' | 'max' | None (default 1)) -- Number of final components to compute. If int, will compute that many. If None, will compute as many as possible. 'min' and 'max' will respectively use the minimum/maximum number of features among views.
regs (float | 'lw' | 'oas' | None, or list, optional (default None)) --
MCCA regularization for each data view, which can be important for high dimensional data. A list will specify for each view separately. If float, must be between 0 and 1 (inclusive).
0 or None : corresponds to SUMCORR-AVGVAR MCCA.
1 : partial least squares SVD (generalizes to more than 2 views)
'lw' : Default
sklearn.covariance.ledoit_wolfregularization'oas' : Default
sklearn.covariance.oasregularization
signal_ranks (int, None or list, optional (default None)) -- The initial signal rank to compute. If None, will compute the full SVD. A list will specify for each view separately.
center (bool, or list (default True)) -- Whether or not to initially mean center the data. A list will specify for each view separately.
i_mcca_method ('auto' | 'svd' | 'gevp' (default 'auto')) -- Whether or not to use the SVD based method (only works with no regularization) or the gevp based method for informative MCCA.
multiview_output (bool, optional (default True)) -- If True, the
.transformmethod returns one dataset per view. Otherwise, it returns one dataset, of shape (n_samples, n_components)
- means_¶
The means of each view, each of shape (n_features,)
- Type
list of numpy.ndarray
- loadings_¶
The loadings for each view used to project new data, each of shape (n_features_b, n_components).
- Type
list of numpy.ndarray
- common_score_norms_¶
Column norms of the sum of the fitted view scores. Used for projecting new data
- Type
numpy.ndarray, shape (n_components,)
- evals_¶
The generalized eigenvalue problem eigenvalues.
- Type
numpy.ndarray, shape (n_components,)
See also
References
- 3
Kettenring, J. R., "Canonical Analysis of Several Sets of Variables." Biometrika, 58:433-451, (1971)
- 4
Tenenhaus, A., et al. "Regularized generalized canonical correlation analysis." Psychometrika, 76:257–284, 2011
Examples
>>> from mvlearn.embed import MCCA >>> X1 = [[0, 0, 1], [1, 0, 0], [2, 2, 2], [3, 5, 4]] >>> X2 = [[0.1, -0.2], [0.9, 1.1], [6.2, 5.9], [11.9, 12.3]] >>> X3 = [[0, 1, 0], [1, 9, 0], [4, 3, 3,], [12, 8, 10]] >>> mcca = MCCA() >>> mcca.fit([X1, X2, X3]) MCCA() >>> Xs_scores = mcca.transform([X1, X2, X3])
Kernel MCCA¶
- class mvlearn.embed.KMCCA(n_components=1, kernel='linear', kernel_params={}, regs=None, signal_ranks=None, sval_thresh=0.001, diag_mode='A', center=True, filter_params=False, n_jobs=None, multiview_output=True, pgso=False, tol=0.1)[source]¶
Kernel multi-view canonical correlation analysis.
- Parameters
n_components (int, None (default 1)) -- Number of components to compute. If None, will use the number of features.
kernel (str, callable, or list (default 'linear')) -- The kernel function to use. This is the metric argument to
sklearn.metrics.pairwise.pairwise_kernels. A list will specify for each view separately.kernel_params (dict, or list (default {})) -- Key word arguments to
sklearn.metrics.pairwise.pairwise_kernels. A list will specify for each view separately.regs (float, None, or list, optional (default None)) -- None equates to 0. Floats are nonnegative. The value is used to regularize singular values in each view based on diag_mode A list will specify the method for each view separately.
signal_ranks (int, None, or list, optional (default None)) -- Largest SVD rank to compute for each view. If None, the full rank decomposition will be used. A list will specify for each view separately.
sval_thresh (float, or list (default 1e-3)) -- For each view we throw out singular values of (1/n)K, the gram matrix scaled by n_samples, below this threshold. A non-zero value deals with singular gram matrices.
diag_mode ('A' | 'B' | 'C' (default 'A')) --
Method of regularizing singular values s with regularization parameter r
center (bool, or list (default True)) -- Whether or not to initially mean center the data. A list will specify for each view separately.
filter_params (bool (default False)) -- See
sklearn.metrics.pairwise.pairwise_kernelsdocumentation.n_jobs (int, None, optional (default None)) -- Number of jobs to run in parallel when computing kernel matrices. See
sklearn.metrics.pairwise.pairwise_kernelsdocumentation.multiview_output (bool, optional (default True)) -- If True, the
.transformmethod returns one dataset per view. Otherwise, it returns one dataset, of shape (n_samples, n_components)pgso (bool, optional (default False)) -- If True, computes a partial Gram-Schmidt orthogonalization approximation of the kernel matrices to the given tolerance.
tol (float (default 0.1)) -- The minimum matrix trace difference between a kernel matrix and its computed pgso approximation, relative to the kernel trace.
- kernel_col_means_¶
The column means of each gram matrix
- Type
list of numpy.ndarray, shape (n_samples,)
- dual_vars_¶
The loadings for the gram matrix of each view
- Type
numpy.ndarray, shape (n_views, n_samples, n_components)
- common_score_norms_¶
Column norms of the sum of the view scores. Useful for projecting new data
- Type
numpy.ndarray, shape (n_components,)
- evals_¶
The generalized eigenvalue problem eigenvalues.
- Type
numpy.ndarray, shape (n_components,)
- Xs_¶
Xs[i] shape (n_samples, n_features_i)
The original data matrices for use in kernel matrix computation during calls to
.transform.- Type
list of numpy.ndarray, length (n_views,)
- pgso_Ls_¶
pgso_Ls_[i] shape (n_samples, rank_i)
The Gram-Schmidt approximations of the kernel matrices
- Type
list of numpy.ndarray, length (n_views,)
- pgso_norms_¶
pgso_norms_[i] shape (rank_i,)
The maximum norms found during the Gram-Schmidt procedure, descending
- Type
list of numpy.ndarray, length (n_views,)
- pgso_idxs_¶
pgso_idxs_[i] shape (rank_i,)
The sample indices of the maximum norms
- Type
list of numpy.ndarray, length (n_views,)
- pgso_Xs_¶
pgso_Xs_[i] shape (rank_i, n_features)
The samples with indices saved in pgso_idxs_, sorted by pgso_norms_
- Type
list of numpy.ndarray, length (n_views,)
- pgso_ranks_¶
The ranks of the partial Gram-Schmidt results for each view.
- Type
list, length (n_views,)
Notes
Traditional CCA aims to find useful projections of features in each view of data, computing a weighted sum, but may not extract useful descriptors of the data because of its linearity. KMCCA offers an alternative solution by first projecting the data onto a higher dimensional feature space.
\[\phi: \mathbf{x} = (x_1,...,x_m) \mapsto \phi(\mathbf{x}) = (z_1,...,z_N), (m << N)\]before performing CCA in the new feature space.
Kernels are effectively distance functions that compute inner products in the higher dimensional feature space, a method known as the kernel trick. A kernel function K, such that for all \(\mathbf{x}, \mathbf{z} \in X\)
\[K(\mathbf{x}, \mathbf{z}) = \langle\phi(\mathbf{x}) \cdot \phi(\mathbf{z})\rangle.\]The kernel matrix \(K_i\) has entries computed from the kernel function. Using the kernel trick, loadings of the kernel matrix (dual_vars_) are solved for rather than of the features from \(\phi\).
Kernel matrices grow exponentially with the size of data. They not only have to store \(n^2\) elements, but also face the complexity of matrix eigenvalue problems. In a Cholesky decomposition a positive definite matrix K is decomposed to a lower triangular matrix \(L\) : \(K = LL'\).
The dual partial Gram-Schmidt orthogonalization (PSGO) is equivalent to the Incomplete Cholesky Decomposition (ICD) which looks for a low rank approximation of \(L\), reducing the cost of operations of the matrix such that \(\frac{1}{\sum_i K_{ii}} tr(K - LL^T) \leq tol\).
A PSGO tolerance yielding rank \(m\) leads to storage requirements of \(O(mn)\) instead of \(O(n^2)\) and becomes \(O(nm^2)\) instead of \(O(n^3)\) 7.
See also
References
- 5
Hardoon D., et al. "Canonical Correlation Analysis: An Overview with Application to Learning Methods", Neural Computation, Volume 16 (12), pp 2639-2664, 2004.
- 6
Bach, F. and Jordan, M. "Kernel Independent Component Analysis." Journal of Machine Learning Research, 3:1-48, 2002.
- 7(1,2)
Kuss, M. and Graepel, T.. "The Geometry of Kernel Canonical Correlation Analysis." MPI Technical Report, 108. (2003).
Examples
>>> from mvlearn.embed import KMCCA >>> X1 = [[0, 0, 1], [1, 0, 0], [2, 2, 2], [3, 5, 4]] >>> X2 = [[0.1, -0.2], [0.9, 1.1], [6.2, 5.9], [11.9, 12.3]] >>> X3 = [[0, 1, 0], [1, 9, 0], [4, 3, 3,], [12, 8, 10]] >>> kmcca = KMCCA() >>> kmcca.fit([X1, X2, X3]) KMCCA() >>> Xs_scores = kmcca.transform([X1, X2, X3])
Generalized Canonical Correlation Analysis (GCCA)¶
- class mvlearn.embed.GCCA(n_components=None, fraction_var=None, sv_tolerance=None, n_elbows=2, tall=False, max_rank=False, n_jobs=None)[source]¶
An implementation of Generalized Canonical Correlation Analysis 8 suitable for cases where the number of features exceeds the number of samples by first applying single view dimensionality reduction. Computes individual projections into a common subspace such that the correlations between pairwise projections are minimized (ie. maximize pairwise correlation). An important note: this is applicable to any number of views, not just two.
- Parameters
n_components (int (positive), optional, default=None) -- If
self.sv_tolerance=None, selects the number of SVD components to keep for each view. If none, another selection method is used.fraction_var (float, default=None) -- If
self.sv_tolerance=None, andself.n_components=None, selects the number of SVD components to keep for each view by capturing enough of the variance. If none, another selection method is used.sv_tolerance (float, optional, default=None) -- Selects the number of SVD components to keep for each view by thresholding singular values. If none, another selection method is used.
n_elbows (int, optional, default: 2) -- If
self.fraction_var=None,self.sv_tolerance=None, andself.n_components=None, then compute the optimal embedding dimension usingutils.select_dimension(). Otherwise, ignored.tall (boolean, default=False) -- Set to true if n_samples > n_features, speeds up SVD
max_rank (boolean, default=False) -- If true, sets the rank of the common latent space as the maximum rank of the individual spaces. If false, uses the minimum individual rank.
n_jobs (int (positive), default=None) -- The number of jobs to run in parallel when computing the SVDs for each view in fit and partial_fit. None means 1 job, -1 means using all processors.
- projection_mats_¶
A projection matrix for each view, from the given space to the latent space
- Type
list of arrays
- ranks_¶
Number of left singular vectors kept for each view during the first SVD
- Type
list of ints
Notes
Consider two views \(X_1\) and \(X_2\). Canonical Correlation Analysis seeks to find vectors \(a_1\) and \(a_2\) to maximize the correlation \(X_1 a_1\) and \(X_2 a_2\), expanded below.
\[\left(\frac{a_1^TC_{12}a_2} {\sqrt{a_1^TC_{11}a_1a_2^TC_{22}a_2}} \right)\]where \(C_{11}\), \(C_{22}\), and \(C_{12}\) are respectively the view 1, view 2, and between view covariance matrix estimates. GCCA maximizes the sum of these correlations across all pairwise views and computes a set of linearly independent components. This specific algorithm first applies principal component analysis (PCA) independently to each view and then aligns the most informative projections to find correlated and informative subspaces. Parameters that control the embedding dimension apply to the PCA step. The dimension of each aligned subspace is the maximum or minimum of the individual dimensions, per the max_ranks parameter. Using the maximum will capture the most information from all views but also noise from some views. Using the minimum will better remove noise dimensions but at the cost of information from some views.
References
- 8
B. Afshin-Pour, G.A. Hossein-Zadeh, S.C. Strother, H. Soltanian-Zadeh. "Enhancing reproducibility of fMRI statistical maps using generalized canonical correlation analysis in NPAIRS framework." Neuroimage, volume 60, pp. 1970-1981, 2012
Examples
>>> from mvlearn.datasets import load_UCImultifeature >>> from mvlearn.embed import GCCA >>> # Load full dataset, labels not needed >>> Xs, _ = load_UCImultifeature() >>> gcca = GCCA(fraction_var = 0.9) >>> # Transform the first 5 views >>> Xs_latents = gcca.fit_transform(Xs[:5]) >>> print([X.shape[1] for X in Xs_latents]) [9, 9, 9, 9, 9]
Deep Canonical Correlation Analysis (DCCA)¶
- class mvlearn.embed.DCCA(input_size1=None, input_size2=None, n_components=2, layer_sizes1=None, layer_sizes2=None, use_all_singular_values=False, device=device(type='cpu'), epoch_num=200, batch_size=800, learning_rate=0.001, reg_par=1e-05, tolerance=0.001, print_train_log_info=False)[source]¶
An implementation of Deep Canonical Correlation Analysis 9 with PyTorch. It computes projections into a common subspace in order to maximize the correlation between pairwise projections into the subspace from two views of data. To obtain these projections, two fully connected deep networks are trained to initially transform the two views of data. Then, the transformed data is projected using linear CCA. This can be thought of as training a kernel for each view that initially acts on the data before projection. The networks are trained to maximize the ability of the linear CCA to maximize the correlation between the final dimensions.
- Parameters
input_size1 (int (positive)) -- The dimensionality of the input vectors in view 1.
input_size2 (int (positive)) -- The dimensionality of the input vectors in view 2.
n_components (int (positive), default=2) -- The output dimensionality of the correlated projections. The deep network wil transform the data to this size. Must satisfy:
n_components<= max(layer_sizes1[-1], layer_sizes2[-1]).layer_sizes1 (list of ints, default=None) -- The sizes of the layers of the deep network applied to view 1 before CCA. For example, if the input dimensionality is 256, and there is one hidden layer with 1024 units and the output dimensionality is 100 before applying CCA, layer_sizes1=[1024, 100]. If
None, set to [1000,self.n_components_].layer_sizes2 (list of ints, default=None) -- The sizes of the layers of the deep network applied to view 2 before CCA. Does not need to have the same hidden layer architecture as layer_sizes1, but the final dimensionality must be the same. If
None, set to [1000,self.n_components_].use_all_singular_values (boolean (default=False)) -- Whether or not to use all the singular values in the CCA computation to calculate the loss. If False, only the top
n_componentssingular values are used.device (string, default='cpu') -- The torch device for processing. Can be used with a GPU if available.
epoch_num (int (positive), default=200) -- The max number of epochs to train the deep networks.
batch_size (int (positive), default=800) -- Batch size for training the deep networks.
learning_rate (float (positive), default=1e-3) -- Learning rate for training the deep networks.
reg_par (float (positive), default=1e-5) -- Weight decay parameter used in the RMSprop optimizer.
tolerance (float, (positive), default=1e-2) -- Threshold difference between successive iteration losses to define convergence and stop training.
print_train_log_info (boolean, default=False) -- If
True, the training loss at each epoch will be printed to the console when DCCA.fit() is called.
- n_components_¶
The output dimensionality of the correlated projections. The deep network wil transform the data to this size. If not specified, will be set to 2.
- Type
int (positive)
- layer_sizes1_¶
The sizes of the layers of the deep network applied to view 1 before CCA. For example, if the input dimensionality is 256, and there is one hidden layer with 1024 units and the output dimensionality is 100 before applying CCA, layer_sizes1=[1024, 100].
- Type
list of ints
- layer_sizes2_¶
The sizes of the layers of the deep network applied to view 2 before CCA. Does not need to have the same hidden layer architecture as layer_sizes1, but the final dimensionality must be the same.
- Type
list of ints
- device_¶
The torch device for processing.
- Type
string
- deep_model_¶
2 view Deep CCA object used to transform 2 views of data together.
- Type
DeepPairedNetworksobject
- linear_cca_¶
Linear CCA object used to project final transformations from output of
deep_modelto then_components.- Type
linear_ccaobject
- model_¶
Wrapper around
deep_modelto allow parallelisation.- Type
torch.nn.DataParallel object
- loss_¶
Loss function for
deep_model. Defined as the negative correlation between outputs of transformed views.- Type
cca_lossobject
- optimizer_¶
Optimizer used to train the networks.
- Type
torch.optim.RMSprop object
- Raises
ModuleNotFoundError -- In order to run DCCA, pytorch and other certain optional dependencies must be installed. See the installation page for details.
Notes
Deep Canonical Correlation Analysis is a method of finding highly correlated subspaces for 2 views of data using nonlinear transformations learned by deep networks. It can be thought of as using deep networks to learn the best potentially nonlinear kernels for a variant of kernel CCA.
The networks used for each view in DCCA consist of fully connected linear layers with a sigmoid activation function.
The problem DCCA problem is formulated from 9. Consider two views \(X_1\) and \(X_2\). DCCA seeks to find the parameters for each view, \(\Theta_1\) and \(\Theta_2\), such that they maximize
\[\text{corr}\left(f_1\left(X_1;\Theta_1\right), f_2\left(X_2;\Theta_2\right)\right)\]These parameters are estimated in the deep network by following gradient descent on the input data. Taking \(H_1, H_2 \in R^{o \times m}\) to be the outputs of the deep network in each column for the input data of size \(m\). Take the centered matrix \(\bar{H}_1 = H_1-\frac{1}{m}H_1{1}\), and \(\bar{H}_2 = H_2-\frac{1}{m}H_2{1}\). Then, define
\[\begin{split}\begin{align*} \hat{\Sigma}_{12} &= \frac{1}{m-1}\bar{H}_1\bar{H}_2^T \\ \hat{\Sigma}_{11} &= \frac{1}{m-1}\bar{H}_1\bar{H}_1^T + r_1I \\ \hat{\Sigma}_{22} &= \frac{1}{m-1}\bar{H}_2\bar{H}_2^T + r_2I \end{align*}\end{split}\]Where \(r_1\) and \(r_2\) are regularization constants \(>0\) so the matrices are guaranteed to be positive definite.
The correlation objective function is the sum of the top \(k\) singular values of the matrix \(T\), where
\[T = \hat{\Sigma}_{11}^{-1/2}\hat{\Sigma}_{12}\hat{\Sigma}_{22}^{-1/2}\]Which is the matrix norm of T. Thus, the loss is
\[L(X_1, X2) = -\text{corr}\left(H_1, H_2\right) = -\text{tr}(T^TT)^{1/2}.\]Examples
>>> from mvlearn.embed import DCCA >>> import numpy as np >>> # Exponential data as example of finding good correlation >>> view1 = np.random.normal(loc=2, size=(1000, 75)) >>> view2 = np.exp(view1) >>> view1_test = np.random.normal(loc=2, size=(200, 75)) >>> view2_test = np.exp(view1_test) >>> input_size1, input_size2 = 75, 75 >>> n_components = 2 >>> layer_sizes1 = [1024, 4] >>> layer_sizes2 = [1024, 4] >>> dcca = DCCA(input_size1, input_size2, n_components, layer_sizes1, ... layer_sizes2) >>> dcca = dcca.fit([view1, view2]) >>> outputs = dcca.transform([view1_test, view2_test]) >>> print(outputs[0].shape) (200, 2)
References
Multiview Multidimensional Scaling¶
- class mvlearn.embed.MVMDS(n_components=2, num_iter=15, dissimilarity='euclidean')[source]¶
An implementation of Classical Multiview Multidimensional Scaling for jointly reducing the dimensions of multiple views of data 10. A Euclidean distance matrix is created for each view, double centered, and the k largest common eigenvectors between the matrices are found based on the stepwise estimation of common principal components. Using these common principal components, the views are jointly reduced and a single view of k-dimensions is returned.
MVMDS is often a better alternative to PCA for multi-view data. See the
tutorialsin the documentation.- Parameters
n_components (int (positive), default=2) -- Represents the number of components that the user would like to be returned from the algorithm. This value must be greater than 0 and less than the number of samples within each view.
num_iter (int (positive), default=15) -- Number of iterations stepwise estimation goes through.
dissimilarity ({'euclidean', 'precomputed'}, default='euclidean') --
Dissimilarity measure to use:
'euclidean': Pairwise Euclidean distances between points in the dataset.
'precomputed': Xs is treated as pre-computed dissimilarity matrices.
- components_¶
Joint transformed MVMDS components of the input views.
- Type
numpy.ndarray, shape(n_samples, n_components)
Notes
Classical Multiview Multidimensional Scaling can be broken down into two steps. The first step involves calculating the Euclidean Distance matrices, \(Z_i\), for each of the \(k\) views and double-centering these matrices through the following calculations:
\[\Sigma_{i}=-\frac{1}{2}J_iZ_iJ_i\]\[\text{where }J_i=I_i-{\frac {1}{n}}\mathbb{1}\mathbb{1}^T\]The second step involves finding the common principal components of the \(\Sigma\) matrices. These can be thought of as multiview generalizations of the principal components found in principal component analysis (PCA) given several covariance matrices. The central hypothesis of the common principal component model states that given k normal populations (views), their \(p\) x \(p\) covariance matrices \(\Sigma_{i}\), for \(i = 1,2,...,k\) are simultaneously diagonalizable as:
\[\Sigma_{i} = QD_i^2Q^T\]where \(Q\) is the common \(p\) x \(p\) orthogonal matrix and \(D_i^2\) are positive \(p\) x \(p\) diagonal matrices. The \(Q\) matrix contains all the common principal components. The common principal component, \(q_j\), is found by solving the minimization problem:
\[\text{Minimize} \sum_{i=1}^{k}n_ilog(q_j^TS_iq_j)\]\[\text{Subject to } q_j^Tq_j = 1\]where \(n_i\) represent the degrees of freedom and \(S_i\) represent sample covariance matrices.
This class does not support
MVMDS.transform()due to the iterative nature of the algorithm and the fact that the transformation is done during iterative fitting. UseMVMDS.fit_transform()to do both fitting and transforming at once.Examples
>>> from mvlearn.embed import MVMDS >>> from mvlearn.datasets import load_UCImultifeature >>> Xs, _ = load_UCImultifeature() >>> print(len(Xs)) # number of samples in each view 6 >>> print(Xs[0].shape) # number of samples in each view (2000, 76) >>> mvmds = MVMDS(n_components=5) >>> Xs_reduced = mvmds.fit_transform(Xs) >>> print(Xs_reduced.shape) (2000, 5)
References
- 10
Trendafilov, Nickolay T. “Stepwise Estimation of Common Principal Components.” Computational Statistics & Data Analysis, 54(12):3446–3457, 2010
- 11
Kanaan-Izquierdo, Samir, et al. "Multiview: a software package for multiview pattern recognition methods." Bioinformatics, 35(16):2877–2879, 2019
Split Autoencoder¶
- class mvlearn.embed.SplitAE(hidden_size=64, num_hidden_layers=2, embed_size=20, training_epochs=10, batch_size=16, learning_rate=0.001, print_info=False, print_graph=True)[source]¶
Implements an autoencoder that creates an embedding of a view View1 and from that embedding reconstructs View1 and another view View2, as described in 12.
- Parameters
hidden_size (int (default=64)) -- number of nodes in the hidden layers
num_hidden_layers (int (default=2)) -- number of hidden layers in each encoder or decoder net
embed_size (int (default=20)) -- size of the bottleneck vector in the autoencoder
training_epochs (int (default=10)) -- how many times the network trains on the full dataset
batch_size (int (default=16):) -- batch size while training the network
learning_rate (float (default=0.001)) -- learning rate of the Adam optimizer
print_info (bool (default=False)) -- whether or not to print errors as the network trains.
print_graph (bool (default=True)) -- whether or not to graph training loss
- view1_encoder_¶
the View1 embedding network as a PyTorch module
- Type
torch.nn.Module
- view1_decoder_¶
the View1 decoding network as a PyTorch module
- Type
torch.nn.Module
- view2_decoder_¶
the View2 decoding network as a PyTorch module
- Type
torch.nn.Module
- Raises
ModuleNotFoundError -- In order to run SplitAE, pytorch and other certain optional dependencies must be installed. See the installation page for details.
Notes
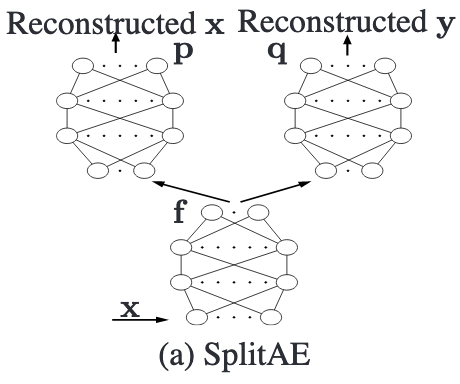
In this figure \(\textbf{x}\) is View1 and \(\textbf{y}\) is View2
Each encoder / decoder network is a fully connected neural net with paramater count equal to:
\[\left(\text{input_size} + \text{embed_size}\right) \cdot \text{hidden_size} + \sum_{1}^{\text{num_hidden_layers}-1}\text{hidden_size}^2\]Where \(\text{input_size}\) is the number of features in View1 or View2.
The loss that is reduced via gradient descent is:
\[J = \left(p(f(\textbf{x})) - \textbf{x}\right)^2 + \left(q(f(\textbf{x})) - \textbf{y}\right)^2\]Where \(f\) is the encoder, \(p\) and \(q\) are the decoders, \(\textbf{x}\) is View1, and \(\textbf{y}\) is View2.
References
- 12
Wang, Weiran, et al. "On Deep Multi-View Representation Learning." In Proceedings of the 32nd International Conference on Machine Learning, 37:1083-1092, 2015.
For more extensive examples, see the
tutorialsfor SplitAE in this documentation.
DCCA Utilities¶
- class mvlearn.embed.linear_cca[source]¶
Implementation of linear CCA to act on the output of the deep networks in DCCA.
Consider two views \(X_1\) and \(X_2\). Canonical Correlation Analysis seeks to find vectors \(a_1\) and \(a_2\) to maximize the correlation between \(X_1 a_1\) and \(X_2 a_2\).
- class mvlearn.embed.cca_loss(n_components, use_all_singular_values, device)[source]¶
An implementation of the loss function of linear CCA as introduced in the original paper for
DCCA9. Details of how this loss is computed can be found in the paper or in the documentation forDCCA.- Parameters
n_components (int (positive)) -- The output dimensionality of the CCA transformation.
use_all_singular_values (boolean) -- Whether or not to use all the singular values in the loss calculation. If False, only use the top n_components singular values.
device (torch.device object) -- The torch device being used in DCCA.
- use_all_singular_values_¶
Whether or not to use all the singular values in the loss calculation. If False, only use the top
n_componentssingular values.- Type
boolean
- device_¶
The torch device being used in DCCA.
- Type
torch.device object
- class mvlearn.embed.MlpNet(layer_sizes, input_size)[source]¶
Multilayer perceptron implementation for fully connected network. Used by
DCCAfor the fully transformation of a single view before linear CCA. Extends torch.nn.Module.- Parameters
layer_sizes (list of ints) -- The sizes of the layers of the deep network applied to view 1 before CCA. For example, if the input dimensionality is 256, and there is one hidden layer with 1024 units and the output dimensionality is 100 before applying CCA, layer_sizes1=[1024, 100].
input_size (int (positive)) -- The dimensionality of the input vectors to the deep network.
- layers_¶
The layers in the network.
- Type
torch.nn.ModuleList object
Initializes internal Module state, shared by both nn.Module and ScriptModule.
- class mvlearn.embed.DeepPairedNetworks(layer_sizes1, layer_sizes2, input_size1, input_size2, n_components, use_all_singular_values, device=device(type='cpu'))[source]¶
A pair of deep networks for operating on the two views of data. Consists of two
MlpNetobjects for transforming 2 views of data inDCCA. Extends torch.nn.Module.- Parameters
layer_sizes1 (list of ints) -- The sizes of the layers of the deep network applied to view 1 before CCA. For example, if the input dimensionality is 256, and there is one hidden layer with 1024 units and the output dimensionality is 100 before applying CCA, layer_sizes1=[1024, 100].
layer_sizes2 (list of ints) -- The sizes of the layers of the deep network applied to view 2 before CCA. Does not need to have the same hidden layer architecture as layer_sizes1, but the final dimensionality must be the same.
input_size1 (int (positive)) -- The dimensionality of the input vectors in view 1.
input_size2 (int (positive)) -- The dimensionality of the input vectors in view 2.
n_components (int (positive), default=2) -- The output dimensionality of the correlated projections. The deep network will transform the data to this size. If not specified, will be set to 2.
use_all_singular_values (boolean (default=False)) -- Whether or not to use all the singular values in the CCA computation to calculate the loss. If False, only the top
n_componentssingular values are used.device (string, default='cpu') -- The torch device for processing.
- model1_¶
Deep network for view 1 transformation.
- Type
MlpNetobject
- model2_¶
Deep network for view 2 transformation.
- Type
MlpNetobject
- loss_¶
Loss function for the 2 view DCCA.
- Type
cca_lossobject
Initializes internal Module state, shared by both nn.Module and ScriptModule.
Dimension Selection¶
- mvlearn.embed.select_dimension(X, n_components=None, n_elbows=2, threshold=None, return_likelihoods=False)[source]¶
Generates profile likelihood from array based on Zhu and Godsie method 14. Elbows correspond to the optimal embedding dimension.
- Parameters
X (1d or 2d array-like) -- Input array generate profile likelihoods for. If 1d-array, it should be sorted in decreasing order. If 2d-array, shape should be (n_samples, n_features).
n_components (int, optional, default: None.) -- Number of components to embed. If None,
n_components = floor(log2(min(n_samples, n_features))). Ignored if X is 1d-array.n_elbows (int, optional, default: 2.) -- Number of likelihood elbows to return. Must be > 1.
threshold (float, int, optional, default: None) -- If given, only consider the singular values that are > threshold. Must be >= 0.
return_likelihoods (bool, optional, default: False) -- If True, returns the all likelihoods associated with each elbow.
- Returns
elbows (list) -- Elbows indicate subsequent optimal embedding dimensions. Number of elbows may be less than n_elbows if there are not enough singular values.
sing_vals (list) -- The singular values associated with each elbow.
likelihoods (list of array-like) -- Array of likelihoods of the corresponding to each elbow. Only returned if return_likelihoods is True.
References
- 13
Code from the https://github.com/neurodata/graspy package, reproduced and shared with permission.
- 14
Zhu, M. and Ghodsi, A., "Automatic dimensionality selection from the scree plot via the use of profile likelihood. Computational Statistics & Data Analysis." 51(2):918-930, 2006